Recently, Associate Professor Li Wang's research group at our institution reported the development of two ultrasensitive electrochemiluminescence (ECL) sensors for miRNA detection. These findings were published under the titles “A tandem DNA nanomachines-supported electrochemiluminescence assay for attomolar detection of miRNA at ambient temperature” and “Poly(3,4-ethylenedioxythiophene):poly(styrene sulfonate)-modified metal-organic frameworks enhancing carbon dots electrochemiluminescence for sensitive miRNA detection”. The papers were respectively published in the Chemical Engineering Journal (a top-tier journal in the fields of engineering, technology, and chemical engineering) and Biosensors and Bioelectronics (a high-impact journal in the field of electrochemistry and biosensors, ranked in the first quartile, Q1). The articles can be accessed at https://doi.org/10.1016/j.cej.2023.148161 and https://doi.org/10.1016/j.bios.2024.116015. Graduate students Jiaxin Peng and Honghong Wang from the class of 2021 are the first authors of these papers.
Method design based on cascade DNA nanomachine:
A novel ultrasensitive, rapid, and straightforward method for detecting miRNA-155 has been developed by integrating tandem DNA nanomachines (DNA Walker and DNA rolling machine) with electrochemiluminescence (ECL) technology. The strategy involves the design of DNAzyme-driven DNA Walkers and catalytic hairpin assembly (CHA)-driven DNA rolling machines on DNA-functionalized electrodes. Detection begins with amplification through the DNA Walker, where the target strand displaces the activated DNAzyme, which sequentially cleaves the phosphodiester bonds downstream (in the 3' direction) of the substrate chain on AuNPs, resulting in the formation of spherical nucleic acids (SNAs). These SNAs then roll along the electrode surface, triggered by the DNA hairpin track, initiating the CHA reaction and enabling secondary signal amplification and ECL signal readout. Quantification of miRNA-155 is achieved by measuring the ECL intensity. Notably, the signal amplification process of the tandem DNA nanomachines, driven by DNAzyme cleavage and CHA, occurs without the involvement of additional enzymes, and the entire process is operational at ambient temperature. Compared to existing DNA nanomachine-based detection methods, this approach offers two significant advantages: (a) simplicity and rapidity, as the entire detection process—comprising only incubation and centrifugation steps—can be completed at room temperature in approximately 70 minutes; and (b) high sensitivity, with a detection limit for miRNA-155 as low as 0.33 aM.
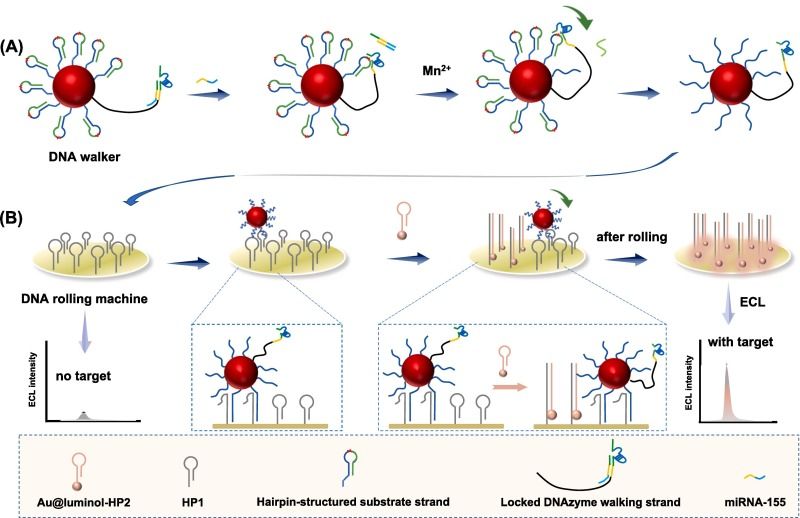
Principle of tandem DNA nanomachine-supported ECL detection of miRNA-155
Design of detection method for CDs/PEDOT:PSS/ZIF-8/AuNPs nanomaterials as ECL emitters:
In this study, PEDOT:PSS-modified CDs/ZIF-8 was synthesised as an efficient ECL emitter, and the novel emitter possessed the following properties: a) the topological order and porous structure of ZIF-8 allowed the CDs to be localised and uniformly distributed within the framework; and b) highly conductive PEDOT:PSS and AuNPs facilitated the electron transfer between the nanomaterials and the electrodes to promote the adsorbed electrochemical activation of CDs in the ZIF-8 framework. As a result, the prepared CDs/PEDOT:PSS/ZIF-8/AuNPs showed 10-fold higher ECL emission than that of individual CDs. Based on the excellent ECL performance and easy labelling of CDs/PEDOT:PSS/ZIF-8/AuNPs, a novel ECL biosensor was successfully constructed by combining the novel composites with DNAzyme-based double-cycling target amplification, achieving an ultrasensitive and highly selective determination of miRNA-21 with a low LOD of 50 aM.
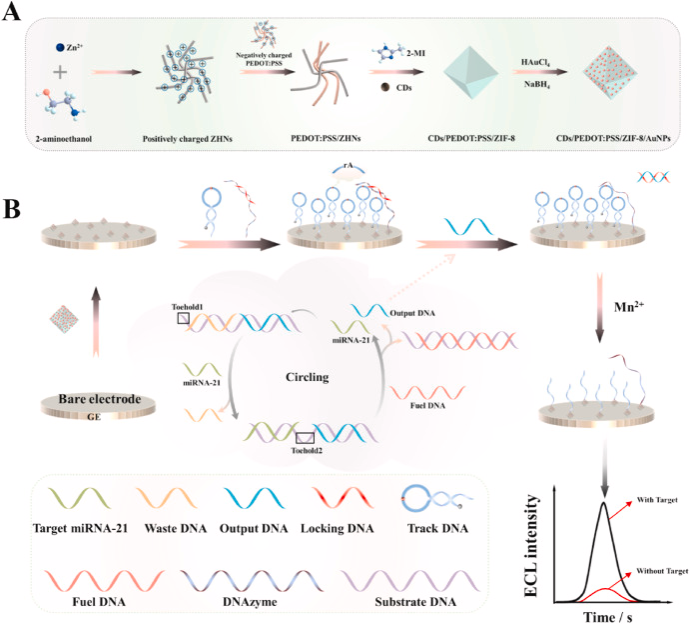
(A)Preparation process of CDs/PEDOT:PSS/ZIF-8/AuNPs. (B) Schematic diagram of the ECL biosensor for miRNA-21 detection.
Overall, two ultrasensitive electrochemiluminescent biosensors were constructed for amplification-free miRNA detection. The LODs for detecting miRNA-155 and miRNA-21 were 0.33 aM and 50 aM, respectively.